St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Childhood Solid Tumor Network moves to the cloud with additional data and analytic tools
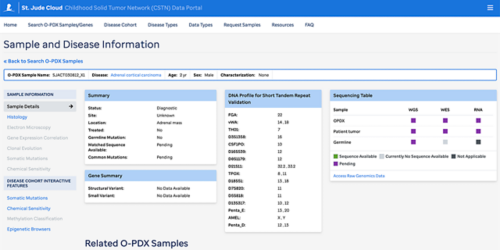
The Childhood Solid Tumor Network data portal has in-depth information on 21 different pediatric solid tumors.
The Childhood Solid Tumor Network, the world’s largest and most comprehensive collection of scientific resources for studying pediatric solid tumors and their biology, has a home on St. Jude Cloud.
“The interactive data portal is a one-stop shop for scientists who are interested in studying pediatric solid tumors,” said Michael Dyer, PhD, chair of the St. Jude Developmental Neurobiology department and a Howard Hughes Medical Institute investigator. “The browser brings the data closer to researchers. The goal is to provide models and share the related data in order to speed cures and better understand the disease.”
Sharing comprehensive patient-derived pediatric cancer information
The data portal comes seven years after Dyer and Alberto Pappo, MD, of St. Jude Oncology, launched the Childhood Solid Tumor Network with support from St. Jude Children’s Research Hospital and Howard Hughes Medical Institute. The goal was to accelerate discoveries related to pediatric solid tumors where patient outcomes have remained largely unchanged for three decades. The resource is available to academic researchers at no charge and with no obligation to collaborate.
The network offers in-depth information on 170 patient-derived samples representing 21 different pediatric solid tumors. A variety of visualization and analytic tools are also available. The tumor types include neuroblastoma, rhabdomyosarcoma and rare tumors. The samples originated from tumor samples donated by patients and families to create orthotopic xenografts. The samples were implanted and grown in the corresponding location in laboratory models.
Pediatric solid tumors are developmental tumors. To understand how tumors begin, grow and spread, they need to be studied in the environment in which they develop. “Not all rhabdomyosarcomas or neuroblastomas are the same,” Pappo said. “The beauty of this resource is that it includes specific subtypes of the different cancers.”
Data and analytic tools available on the CSTN Data Portal include:
- Next-generation, whole-genome sequencing of the patient tumors, germline and the orthotopic patient-derived xenografts (O-PDX). These are presented as an interactive application, allowing for customized heatmaps to represent mutations and individual gene searches. Raw genomic sequences are also uploaded and available.
- Interactive browsers for epigenetic and, in some cases, proteomic data
- Histology images with tissue-specific stains and electron microscopy images at three magnifications for each sample
- Clonal analysis of the matched patient samples and orthotopic xenografts
- Sample-level search and visualization of specific characterization data that let users filter by factors such as patient’s age at diagnosis or primary versus recurrent tumors
- Preclinical pharmacokinetic reports, tumor propagation protocols and more
What is the future of the CSTN Data Portal?
Single-cell RNA sequencing of patient and O-PDX is underway. Additional tumor samples and data are added to the network as they become available.
Academic researchers can also request cryopreserved cells from O-PDX, as well as fresh frozen tissue or cells and formalin-fixed paraffin-embedded tissue blocks. Request resources.
Visit the Childhood Solid Tumor Network Data Portal.






