St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
What you’re missing if you’re only sequencing exomes or RNA
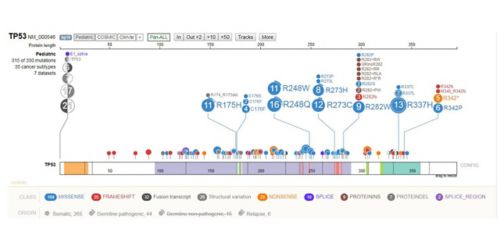
Visualization from St. Jude Cloud, which offers data from the Genomes for Kids clinical trial, to be highlighted at the 2018 meeting of the American Society for Human Genetics.
A genome is a complicated place, and a cancer genome is even more complicated. Finding the key mutations driving a cancer poses an enormous challenge. Next-generation sequencing technology makes the job feasible, though analyzing exomes or transcriptomes is more common than using whole-genome sequencing (WGS)—mainly because WGS takes longer and costs more.
But what is the potential human cost of omitting WGS from the picture? New findings suggest many actionable mutations are missed without WGS in the mix.
The findings come from a St. Jude-led clinical trial called Genomes for Kids, which sequenced tumor and matched germline samples for 253 pediatric cancer patients using three platforms: RNA-Seq, whole-exome sequencing (WES) and WGS.
“To our knowledge, this is the first clinical study where this comprehensive three-platform genomic sequencing approach was offered prospectively to all pediatric oncology patients,” said Kim Nichols, MD, the study’s principal investigator.
For nearly 80% of cases in the study, this comprehensive approach yielded at least one finding that could be used to guide care.
The results will be presented this week at the annual meeting of the American Society for Human Genetics (ASHG) in San Diego. (View the abstracts on these results and findings from the trial related to cancer predisposition.)
St. Jude Progress spoke with Nichols, director of St. Jude Cancer Predisposition; study co-PI Jinghui Zhang, PhD, chair of St. Jude Computational Biology; and Scott Newman, PhD, who leads the clinical genome analysis team within St. Jude Computational Biology. All three were centrally involved in the work.

The study, which involved a collaborative effort across disciplines, showed the comprehensive three-platform approach yields clinical findings that could be used to guide care. From left: Kim Nichols, MD, director of St. Jude Cancer Predisposition, James R. Downing, MD, St. Jude CEO and President, and study co-PI Jinghui Zhang, PhD, chair of St. Jude Computational Biology.
St. Jude Progress: Tell me about the three-platform sequencing approach. Why is it better?
Zhang: Different sequencing methods are better for identifying different alterations. It isn’t surprising that WGS discovered high-risk mutations that WES and RNA-Seq did not.
Newman: And doing RNA, exome and whole-genome sequencing allowed us to cross-validate the variants we found across all three platforms. That got us high-accuracy calls with a low limit of detection.
SJP: What findings have been most exciting so far?
Newman: There are lots of exciting ones to list. From my end, it’s the findings you can’t get by any other technology – ones you can’t get by gene panel sequencing, or locus-specific tests. We found a few really interesting ones.
For example, there was a neuroepithelial tumor that had been described by investigators at St. Jude. Coming into the study we thought it was a high-grade glioma, but it was the wrong diagnosis. We found a really interesting structural rearrangement of a gene called MN1, which meant the tumor was probably part of an entirely new group discovered only a couple of years ago.
It would have been very hard to find this event without doing WGS and RNA-Seq and discussing it in a committee. This was a structural rearrangement in a region that was not well understood and not easy to design targeted assays for. The only way to get to it was through a WGS approach.
Zhang: RNA-Seq is also very important. It helps verify the expression status of oncogenic variants, and detect fusion genes, which is very important in pediatric cancer. Part of what we are presenting at ASHG is our finding of novel fusions that were not previously known.
SJP: What’s the turnaround time for the results – is it short enough to inform clinical decisions?
Newman: Yes, and that’s a key point. We hope to drive the wider adoption of WGS as a method to assess pediatric tumors. We also think that simultaneous testing for germline predisposition genes should happen. This study shows it can be done in a clinically relevant timeline. At the end of the study, we were turning it around in 40 days. Our clinical service can now do it even faster.
Zhang: Because of the findings and the ability to turn around in a clinically relevant timeframe, we now have a new study for pediatric leukemia that specifically targets kinase fusions. That requires a turnaround time of two weeks. Genomes for Kids set the stage for this new study, which is designed to use genomic information to determine the treatment options that patients should receive.
SJP: What do you consider actionable results? Did you find a bunch of targetable mutations?
Newman: Genomes for Kids was a feasibility study. We never set out to treat people with directly druggable mutations; we wanted to explore what “actionable” means. Diagnostic mutations are actionable. So are prognostic and targetable mutations. Overall, based on our three-platform sequencing test, 79% of our patients had at least one thing an oncologist would want to know about.

Scott Newman, leader of the clinical genome analysis team in the St. Jude Department of Computational Biology, will present new scientific findings from the Genomes for Kids clinical trial at the ASHG annual meeting in San Diego on Wednesday, Oct. 17, 2018.
SJP: How do you see this approach guiding care?
Nichols: Because so few of the molecular lesions in pediatric cancer are targetable by specific drugs, currently it is the diagnostic and prognostic insights provided by the three-platform approach that appear most clinically impactful. From a diagnostic perspective, tumors may look the same under a microscope, but the identification of specific genetic changes can direct you to the correct diagnosis, and therefore, the most appropriate therapy. From a prognostic perspective, you will have different risk stratifications depending on results.
It’s important to keep in mind that the vast majority newly diagnosed pediatric cancer patients are going to go on a frontline therapeutic trial. In general, they are not going to be pulled off of what are currently considered as gold standard therapies, and treated as a one-off based on their genomic findings.
The analysis of germline variants is key. It can tell us about the risk for additional tumors down the road, risk of therapy-associated toxicities, need for surveillance, and risks for family members.
SJP: How have you started using these findings in the clinic?
Nichols: This feasibility study was relatively small, but clinical genomic sequencing is ongoing at St. Jude. It’s now become a part of our standard of care where tumor testing is offered by primary oncologists to patients with newly diagnosed or relapsed tumors and germline testing is offered by our Cancer Predisposition team to all patients. Each month we identify another 4 to 5 children with an underlying cancer predisposition syndrome. At St. Jude we offer testing and guidance regarding tumor surveillance to other family members as part of our predisposition program.
SJP: So, would you recommend that everyone do three-platform sequencing for their pediatric cancer patients? Does the benefit justify the cost?
Zhang: We would recommend prospective next-generation sequencing (NGS) testing for all pediatric oncology patients, for sure. When we started this study, the cost seemed insurmountable. But now, it is becoming more feasible to give this testing to all children as the cost for sequencing is reduced.
Nichols: Compared with the cost of many other procedures that children with cancer undergo, the cost is likely comparable, or even less – for example, compared with complex surgical procedures or multiple radiology tests.
Zhang: Yes. We love WGS because you do one assay, you get everything. There is more you can learn in the future from the data you have accumulated, instead of ordering one assay at a time.
SJP: How did clinical sequencing get started at St. Jude?
Zhang: It started with the St. Jude—Washington University Pediatric Cancer Genome Project (PCGP). Our CEO, Dr. Jim Downing, was involved in the PCGP, and he’s the one who first defined the strategy of putting all three platforms together for clinical sequencing.
After the PCGP, we conducted our first three-platform clinical pilot study with 78 pediatric patients, which was just published in Nature Communications. Then came Genomes for Kids, which is when we started to work on patients in real time.
SJP: What were some of the biggest challenges in setting up the clinical sequencing pipeline?
Zhang: We had to figure out how to curate data, and how to work the whole setup of multidisciplinary review of cases by a committee. We had to get CLIA certification and CAP accreditation for both the laboratory and the computational pipeline together as part of a package.
Kim’s team determined how to define the consent and enroll the patients, and worked on the genetic cancer predisposition/germline aspects of this study. My team focused on somatic mutations – looking for driver lesions. And we developed novel computational methods in my lab for this clinical program.
Newman: Part of the study was to assess the feasibility of handling all this sequencing. We could end up with hundreds or thousands of these variants and not know what to do with them. So Jinghui’s team designed a way to triage and rank variants in order of importance; we called the approach Medal Ceremony because it was developed during the Olympics.
Actually, it turned out that the reports were gratifyingly short because we were able to easily drill down into important variants. The genome analyst looked at tens to hundreds of mutations per case. The take-home message is: including whole genome sequencing in your assay will not overburden your diagnostic service.
Zhang: Yes. Dr. David Ellison, who is the chair of Pathology, is a co-Investigator on the study, and Dr. Pathology set up the infrastructure and workflow, especially in the clinical service component – making sure samples are acquired early enough to get sequencing and analysis done in a timely fashion relevant for treatment.
SJP: You said you’re sharing the data. Can you tell people where to find it?
Newman: In April, we launched a platform called St. Jude Cloud to distribute data to the research community. The Genomes for Kids data is already there, and people have been using it. This data is not locked away; it’s available to the research community.
There will be a St. Jude Cloud booth in the exhibit hall at ASHG to tell people what data is in there and how they can use it.
Zhang: The data-sharing initiative is very important. One reason we want to share data with the community is so people can see for themselves how powerful WGS is. There are so many things you can learn from the whole genome, and our single institution is not big enough to fully explore the really important genomics signatures embedded in the data.
We hope to involve the whole community – not just people in pediatric cancer, but also adult cancer researchers and computational biologists. All of us in the research community can learn a lot more from the data to help our patients and children around the world.






