St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Spatial Omics
Advances in next-generation sequencing and imaging-based approaches have led to a revolution in our ability to understand cellular function in spatial context.
Spatial Omics technologies
Bedrock technologies that underlie spatial transcriptomics date back to the 1970s. In situ hybridization (ISH) was one of the earliest technologies developed to visualize gene expression in space. The 1980s and early 90s witnessed the emergence of the enhancer and gene trap screen, followed in the late 90s by whole-mount ISH and an array of fluorescent ISH capabilities. The rise of highly multiplexed, high resolution, and quantitative approaches shepherded the creation of gene expression atlases. Advancements in downstream analysis tools and integrated pipelines provide the robust computational methods to decipher spatial organization of cellular properties captured in these data.
Modern-era technologies fall into discrete categories: next generation sequencing-based approaches and imaging-based approaches, distinguished by methods used for data collection.
OVERVIEW
Maintenance of spatial context is crucial for understanding homeostasis and disease as cellular organization is intimately linked to biological function. Pathological changes in gene expression can manifest in specific niches across a particular tissue and emerge dynamically within different cell populations. The omics revolution has expanded our ability to characterize cells through interrogation of the full genome, transcriptome, or proteome. Until recently, these techniques could not be applied in situ, resulting in the loss of spatial relationships.
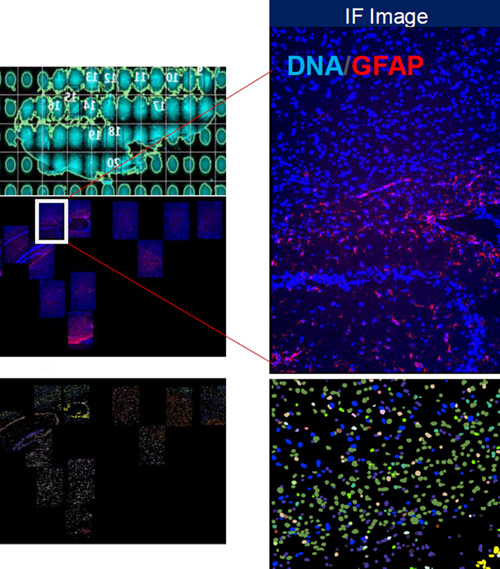
Fluorescence signal (left) and assignment of individual transcripts by cell segmentation (right) via RNA/ISH-based Xenium methodology performed on human breast cancer sample.
IMPACT
Spatial omics offer high-throughput solutions to assess the spatial organization and cell types within niches and understand their intercellular communication. Spatial omics combines next generation seq/high level multiplexing with imaging modalities, which gives depth and clarity to our insights into spatial distribution of gene expression, cell-state transitions, and cell-cell interactions.
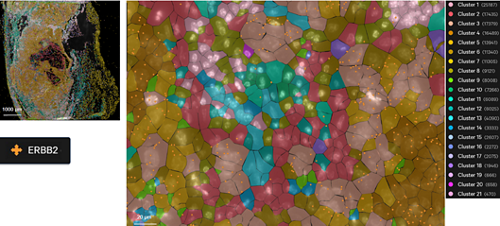
Supervised cell type clustering applied to results generated by RNA/ISH-based CosMx for mouse brain sample.
Technologies
In situ sequencing (ISS) enables direct read-out transcript sequences within a tissue via ligation, gene barcodes, or short fragment cDNAs. RNA is reverse transcribed and amplified by rolling circle amplification, followed by sequencing.
Applications: FISSEQ, StarMAP, etc.
In situ hybridization (ISH) builds on ISS technology, facilitating target sequence detection using fluorescent labeled probes. Sequential rounds of hybridization and imaging combined with barcoding enable substantial multiplexing.
Applications: MERFISH, seqFISH, smFISH, etc.
Though they require a prior knowledge of transcript targets, the throughput of ISS and ISH methods has substantially increased in recent years, and their high resolution facilitates sub-cellular interrogation.
NGS-based methods build on the innovation of single cell RNA sequencing (scRNA-seq) to incorporate spatially barcoded RNAs on microarray slides. These barcodes are used to map spatial position, while traditional sequencing reads map to the genome. These approaches offer the advantage of unbiased interrogation of large regions of tissue.
Applications: DBiT-Seq, HDST, Pixel-seq, Slide-seq, etc.
Multi-omics approaches continue to develop, expanding the focus from the transcriptome to the proteome, epigenome, and metabolome. Targeted co-detection of proteins or DNA genomic loci via high-throughput spatial mapping paired with spatial transcriptome data are starting to reveal snapshots of tissue complexity.
Applications: Multiplexed RNA-protein ISH, MIBI, etc.
The St. Jude Center for Spatial Omics
At St. Jude Children’s Research Hospital, we seamlessly integrate basic, translational, and clinical research to advance cures for catastrophic childhood diseases. Over the past decade, we have led major efforts to profile the genomic landscape of pediatric cancers and integrate those data with transcriptomic, epigenetic, and proteomic data. With recent advances in single-cell profiling using next-generation sequencing platforms, researchers at St. Jude are now elucidating the cellular heterogeneity of pediatric cancer and other experimental models.
Single-cell multi-omic profiling, combined with spatial assay platforms, provides an unprecedented opportunity to elucidate the cellular heterogeneity in normal development, cancer, and other catastrophic childhood diseases. Our strength in advanced microscopy, computational biology, cancer modeling, and the rich resource of tissue samples provide a unique opportunity to accelerate discovery and advance cures.
As part of the 2022-2027 St. Jude Strategic Plan, we are building a new institutional shared resource focused on implementing cutting edge spatial omics platforms. The St. Jude Center for Spatial Omics (CSO) is proud to lead this groundbreaking endeavor. Our experience as part of the Human Tumor Atlas Pilot Project and Human Tumor Atlas Network has provided us with the expertise to rapidly accelerate development of this unique resource to serve all investigators at St. Jude.
Dr. Jasmine Plummer
Dr. Jasmine Plummer is a trained genomicist who brings multi-dimensional expertise to her role as Director of the Center for Spatial Omics. She received her PhD in Molecular Genetics from the University of Toronto and completed a Master’s degree in neuroscience with a speciality in comparative neurobiology and neurodevelopment at Dalhousie University in Nova Scotia, Canada.
Dr. Plummer’s postdoctoral work at Children’s Hospital Los Angeles implemented a systems biology approach to examine the genetic risk of neurodevelopmental disorders. As an Autism Speaks postdoctoral fellow, her research concentrated on the discovery and functional characterization of gene regulatory networks implicated in the development of autism.

Throughout her career, Dr. Plummer has used a multi-omics approach to examine genetic risk as a factor of oncogenesis across cancers, including ovarian, breast, and prostate, and neurodevelopmental disorders.
As Director of the Center for Spatial Omics, Dr. Plummer leverages her extensive research experience and knowledge of data integration, computational pipeline implementation, and program management to lead an omics-centered approach that advances research of pediatric catastrophic diseases.