St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Microscopes: Paths and barriers to discovery and super resolution
Spatial flythrough video through a dual-colored nucleus of a mammalian cerebellar granule neuron. This 3-D rendering illustrates the two chromatin subdomains (blue or copper colors) defined by the overlap of distinct chromatin proteins imaged by cryo-SR/EM. A thin section of electron microscopy data (gray) slices through the nucleus.
The microscope remains our intuitive tool to observe the hidden subcellular details of our body’s organs and cells. The main barrier to microscopic discovery is one simple word: resolution.
But recent advances in microscope technology allow us to see how neurons mature or tumors can begin to form during brain development — and it’s all attributed to simply being able to take a better picture.
Resolution revolution
Light microscopes generally can only resolve 200-nanometer sized objects (~0.3% the width of a human hair). Just imagine, anything below 200-nanometers cannot be accurately seen with conventional microscopes because they are simply too small. The electron microscope, which can resolve objects in the tens of nanometers, helps. But they produce black and white images that don’t reveal molecular details in the photos they provide.
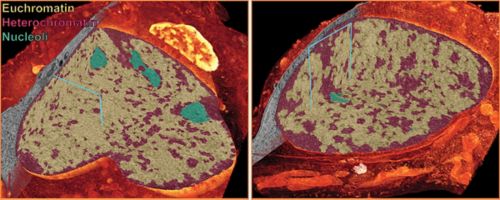
Comparison of the nuclei of a cerebellar neuron progenitor (left) to a differentiated cerebellar granule neuron (right). A cut-away to each 3D rendering of electron microscopy data reveals that the progenitor nucleus is not only much larger than the differentiated neuron, but has a different organization of euchromatin, heterochromatin and nucleoli.
Luckily for biologists, groups of physicists, optics specialists and engineers in the 21st century spearheaded an absolute revolution in the capabilities of both light and electron microscopes. In 2014, Drs. Eric Betzig, Stefan Hell and William Moerner received the Nobel Prize in chemistry for inventing so-called super-resolution microscopy that pushed the resolution of light microscopy to below 20-nanometers. In parallel, electron microscopists have produced ultra-stable new instruments that can section through specimens with such precision that even brain tissue can be reconstructed at 4- or 8-nanometer resolution.
The cryo-SR/EM pipeline
The pace of microscopy innovation led Eric Betzig and Harald Hess from Janelia Research Campus to invent the cryo-SR/EM microscopy described in the journal Science. The pipeline involves imaging cells with super-resolution light microscopy and then imaging the same cells with a next-generation electron microscope. When both image types are computationally combined, the electron microscopy provides an ultra-high-resolution 3-dimensional 4- or 8-nanometer snapshot of every facet of a cell, and the super-resolution light data highlights the location of any protein or cellular machine. Just like astronomers needed the Hubble Space Telescope to peer deep into the universe, cryo-SR/EM microscopy is a powerful tool to peer into the depths of cells for biologists to get their “eureka moments.”
St. Jude and advanced microscopy
So, what do cryo-SR/EM and the microscope revolution have to do with St. Jude Children’s Research Hospital? In 2016, the Developmental Neurobiology (DNB) Department at St. Jude became first in the country to adopt a commercial version of a lattice light sheet microscope (LLSM), another of Eric Betzig’s bleeding-edge microscopy inventions. When Daniel Stabley, DNB’s imaging manager, and I were the first to utilize LLSM to ask a neurobiology question, the unexpected happened: we were soon not only in contact with Eric Betzig but were quickly in the thick of discussions to utilize one of his newly developed microscopes!
What did we do with cryo-SR/EM?
I study how developing neurons mature and modify their cellular architecture to productively assemble into neuronal circuits. This information not only helps us understand how the brain forms during development but reveals pediatric disease processes related to cognitive deficiency and brain tumorigenesis. The name of the game in my field is cellular form instructs function, so we had been urgently looking for techniques like cryo-SR/EM. LLSM imaging showed us how live neurons rapidly repack the DNA in their nucleus as they begin to express new genes during differentiation. Cryo-SR/EM snapshots before and after differentiation went dramatically beyond that to identify the nuclear domains that changed in the process and showed that neurons unexpectedly pack unique classes of genes. These results are an exciting start to understanding how neurons organize gene expression in differentiation during health and disease.
It takes a team
Teamwork was one of the most rewarding parts of the study. Besides Betzig and Hess advising instrument design, their lab members, David Hoffman and Gleb Shtengel, operated cryo-SR/EM instruments with a precision rivaling mission control of a Mars rover mission. Kirby Campbell, a postdoctoral fellow in my lab and Abbas Shirinifard, DNB’s Image Analysis Manager, worked painstakingly with open-source software to fuse microscopy data and torture test analysis pipelines to discover the biology behind the images. Daniel Stabley optimized image reconstruction and pioneered the use of Blender software to render cryo-SR/EM data that was not only beautiful but was essential to reveal biological insight. I guided the logistical and conceptual direction of the studies in neurons plus went to Janelia Research Campus to prepare all of the neurons imaged with cryo-SR/EM. All of this was brought together through 10,000 Slack messages between St. Jude and Janelia!
Success from ... failure?
Some scientists describe how discoveries were made from unexpected results, but few admit that a breakthrough came from failure.
Using cryo-SR/EM was not our first visit to Janelia: the previous year, we went to use an unrelated microscope, and long story short, it was a bit of a flop. On the last day of that visit, after the senior scientists left for the weekend, Daniel Stabley and David Hoffman commiserated over the disappointing results when both realized our lab might also have perfect samples for cryoSR/EM. That started a year-long planning process that was equivalent to setting up a satellite lab for just a week for the successful cryo-SR/EM visit.
The first visit taught us priceless logistical and communication lessons with Janelia that was the foundation of our contribution to the Science paper.






