St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
St. Jude Family of Websites
Explore our cutting edge research, world-class patient care, career opportunities and more.
St. Jude Children's Research Hospital Home

- Fundraising
Chaperones: probing the structural secrets of the cell’s front-line protein protectors
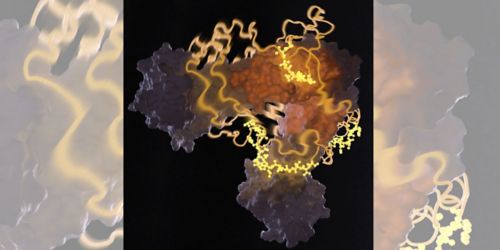
This image from nuclear magnetic resonance spectrometry shows a heat-shock protein bound to an unfolded protein. Charalampos Kalodimos, PhD, and others recently discovered that protector proteins, called chaperones, help ensure protein folding occurs correctly.
Molecules called chaperones are the first line of defense against proteins going “bad” in cells. Normally, proteins are folded from an initial string-like shape into globular functioning enzymes and other cell components. But when a cell is under stress, proteins may unfold, misfold or form into toxic clumps. Such malfunction can give rise to neurodegenerative diseases and other conditions.
Chaperones, found in all cells and all organisms, bind transiently to proteins to prevent such malfunctions. However, until our work, nobody had known the structural details of how a chaperone binds to its “client” protein. Published in Science, our latest work used nuclear magnetic resonance (NMR) spectroscopy to capture the highly transient, unstable molecular interactions between a chaperone and its protein client.
In our Biomolecular NMR Spectroscopy Center, we have just added a 1.1GHZ NMR spectrometer—the world’s most powerful NMR instrument for studying proteins, RNA and other biological molecules. This machine enables us to analyze the structure of molecules that so far have remained beyond our reach.
In NMR spectroscopy, intense magnetic fields are used to tweak a molecule or molecular complex to produce signals that can be analyzed to reveal their structure. NMR spectroscopy can uniquely "see" in great detail the interactions of biological molecules undergoing rapid changes in liquid solution, which is their natural environment. By contrast, other structural techniques, such as X-ray crystallography, can yield detailed structures, but only in static crystallized molecules.
In our experiments, we used NMR spectroscopy to analyze the interaction between a biologically critical three-part chaperone machine called Hsp40-Hsp70-NEF and a protein client. These three linked components interact with the protein to protect it from misfolding or aggregation—which could be catastrophic for the cell.
The Hsp40 molecule interacted with proteins that we biochemically manipulated to keep them in their unfolded state. This intricate molecular interaction—like complex puzzle pieces fitting together—had never before been mapped. Our findings are especially important because this chaperone complex is found in all cells and in all kingdoms of living things.
Once we had this detailed structural information, we tested its validity by selectively altering the Hsp40 molecule by mutation at what we called “hotspots.” These were units of the Hsp40 molecule, called amino acids, that we knew were critical for the chaperone-client interaction. If those hotspot mutations rendered the Hsp40 non-functional, that confirmed that our structural information was correct.
Particularly fascinating about our structural findings was that Hsp40 interacts very differently with the same client proteins than with two other chaperones we previously analyzed. We know the multitude of chaperones in cells come in different sizes and architectures; and now we know they also have different binding mechanisms that lead to varying biological outcomes. We are now analyzing a range of chaperones to discover whether there are common structural themes among them.
Also exciting is our exploration of an important property of the tripartite Hsp40-Hsp70-NEF complex. This chaperone machine is capable not only of protecting its client protein from misfolding, but of actively folding the protein back to its active, or “native” state. Understanding the structural details of this critical process is one of the major goals of our future studies.
We know that disorders including cancer and neurodegenerative diseases are associated with over-production of chaperones or genetic mutation that alters their structure. However, we only now have the ability to study chaperone-client interactions to begin to understand their association. We hope our basic studies will shed light on these mysterious molecules and ultimately lead to clinical treatments.
The St. Jude Structural Biology Department, of which I am chair, has a fully integrated complement of analytical machines to serve our researchers. Besides NMR spectrometry, the department offers techniques such as X-ray crystallography, cryo-electron microscopy and tomography, single-molecule spectroscopy and mass spectrometry. Each of these yields unique pieces of the puzzle of the 3-D structures of biological molecules.






